Karatan Lab
Josiah Zayner
Appalachian State University
Department of Biology
Rankin Science North
319 (Office), 303 (Lab)
828-262-6742

Role of MbaA and NspS in V. cholerae biofilm formation
Vibrio cholerae is a gram negative bacterium and the pathogenic agent of the human disease cholera. If left untreated cholera can kill a person in less than 24 hours. In the past 185 years, V. cholerae has spread nearly globally nine times in a pandemic nature. The persistence of V. cholerae in its natural environments is partially attributed to its ability to form biofilms. The focus of my research is the study of V. cholerae biofilm formation and the protein interactions in the bacterium that regulate this process. By using site directed mutagenesis and immunoprecipitation assays I plan to elucidate the structural and functional aspects of two proteins, MbaA and NspS, involved in the biofilm formation pathway of V. cholerae.
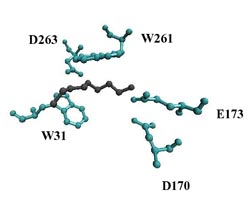
The V. cholerae gene mbaA codes for the inner-membrane protein MbaA. MbaA has been shown to down-regulate biofilm formation and reduce the transcription of exopolysaccharide synthesis (vps) genes, though the exact mechanism is still unclear. The V. cholerae gene nspS codes for the putative norspermidine binding protein NspS. NspS has been implicated as a periplasmic sensor, which binds the inner-membrane protein MbaA and is thought to be activated by the binding of the polyamine, norspermidine. Mutations removing the nspS gene have caused a decrease in biofilm formation and vps gene transcription.
Using mutational analysis followed by biofilm assays and immunoprecipitations, I plan to elucidate the functional characteristics of NspS. Mainly, I am interested in how the various mutations in the proposed norspermidine binding site of NspS affect this protein's role in biofilm formation and its predicted interaction with MbaA.